Key Concepts
DERIVA is a platform for acquiring, organizing, discovering and managing collections of scientific data. Often, the data we are concerned with is in a file. Typically, an investigation will involve many different files and file types. For example, we may have a set of image files, and for each image file, create a spreadsheet or comma-separated value (CSV) file which describes the results of analysis on each image.
Old Way: Ad hoc files and directories
In a world without DERIVA, the investigator would try to manage all of the different files by creating a directory structure and file naming convention for each image/CSV pair. For example, one might give each image and CSV file the same file name, with different extensions (e.g. image1.img and image1.csv).
You might have so many files that it’s hard to find them, or there might be different types of images each with a different extension, so you might create separate directories for the images and CSV so you don’t have to rely on file extensions.
To keep track of where the images came from, you might have a more “meaningful” file name, like mouse26.img, and so on.
If you want to share your data with a collaborator, you need to make sure that they understand your directory and file naming conventions, and if you want to make sure you are looking at the same data, you need to make sure no one updates the files that you are sharing.
It’s not too hard to see how this strategy will become too complex very rapidly.
New Way: Model and manage the relationships between data
DERIVA helps manage the complexity by making it easy to describe the relationships between different files and use those descriptions to help find and share data of interest.
Of course, there are many different types of files one might be interested in, and the relationships between those files might change over time, so DERIVA also makes it easy to adapt to changing data types and relationships.
DERIVA organizes data by specifying an explicit data model using the well known entity/relationship (ER) modeling approach.
REVIEW: (What do we mean by “entity” in DERIVA? How is this different from “asset”?:) In DERIVA, an entity is equivalent to the representation of a single unit of data (or “type of data”?) that will be ingested into a particular DERIVA deployment. The ER model maps out relationships between these entities.
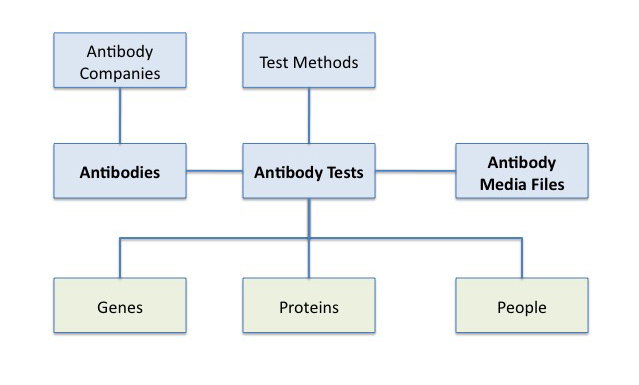
REVIEW: For example, in the (Re)Building A Kidney deployment, one of the types of data shared in its repository are antibodies. The following data model describes the different entities needed for a complete representation of an “Antibody” as well as what other entities are related; e.g., antibody tests have a direct relation to “proteins”.
DERIVA provides mechanisms for:
creating and modifying an ER model,
querying, adding and updating entities that conform to that model.
The DERIVA platform uses model introspection to continually track the current set of relationships between data and to adapt the user and programmatic interfaces to the current model.
DERIVA keeps track of both the data files (or assets) and the model that organizes the data.
Because DERIVA is used to support reproducible science, we generally assume that once an asset is made known to DERIVA (or ingested), its actual contents will not change. To ensure this, DERIVA uses an object store called HATRAC to hold assets. Other object stores such as AWS S3 may also be used but in any case, DERIVA models generally assume that the asset can be referred to uniquely by a URL, and that the contents of the asset will not change. DERIVA usually makes sure this is the case by using checksums on every object.
The data model and instance information (TBD: what does this mean?) about specific entities are stored in our data catalog which is called ERMRest. ERMRest is a catalog service with a web services interface for:
modifying and retrieving the ER model,
creating and modifying new entities, and
specifying policies.
Each ERMRest service is referred to by a URL in the form:
https://mycatalog.isi.edu/ermrest/1
DERIVA allows many catalogs to co-exist and each catalog is referred to by an integer catalog ID, so we might also have:
https://mycatalog.isi.edu/ermrest/2.
The catalog and assets can be accessed directly using a RESTful web services interface. However, to make it easy to use on a daily basis, DERIVA has a powerful, browser-based user interface called Chaise. Chaise is a model-driven user interface (UI), which means that it uses catalog introspection to dynamically adapt the UI to the current data model. Changes to the model, such as adding a new attribute or entity type, are immediately reflected in the user interface.
The Chaise UI is highly configurable to specify how data is presented, which entities should be displayed and in what order, and many other display elements.
To support this customization, Chaise uses an extensible set of annotations that are stored in the catalog along with the data model.
DERIVA also has a programmatic interface written in Python called deriva-py. It is more convenient to use the Python SDK
Assets
One of the primary objectives of Deriva is to manage collections of assets. In Deriva, an asset is a blob, a sequence of bytes with some basic properties:
It can be referred to by a globally unique resource name, i.e. a URL
In general, the sequence will not change without the resource name changing
It will often be characterized by a checksum which is computed from its contents
It will have a known length
It may have a convenient human understandable name
A deriva catalog will generally contain one or more asset tables, which are then orginized into more complex data descriptions to help interpret the different types of assets and figure out how assets relate to one another.
The Catalog Model
TBD: Describe catalog, schema, table, columns, and constraints
Representing entities in a catalog
In DERIVA, an entity is represented by a table in a catalog.
The table has one column for each attribute that describes a characteristic of the entity.
TBD: example of such a table
Representing relationships between entities
Cardinality
One-to-one (1:1) relationships
One-to-many (1:m) relationships
Inbound foreign key constraints
Many-to-one (m:1) relationships
Outbound foreign key constraints
Many-to-many (m:n) relationships
The most general type of relationship we have is a many-to-many (m:n) in which any record A can be related to any number of records B and vice versa.
A common example of this is in relating protocol steps to a sample. Each sample might have multiple steps, while a specific step might be used to prepare more then one sample.
TBD: add real-world examples of this.
In DERIVA, m:n relationships are expressed using an association table, which has two foreign key constraints, one to each of the tables being related. Each row in the association table captures the relationship between one entity in each table. By repeating the same value in the first foreign key, we can have multiple entities in table two associated with a single entity in table one, and visa versa.
TBD: add diagram
To streamline interactions in the DERIVA UI, we try to identify when a table is acting as an association table. For this purpose, we look for pure binary association tables. We say a table is pure binary if:
It only has two foreign keys
Those keys cannot have NULL values
There is a primary key constraint on the two foreign keys; i.e., each pair must be unique.
If those conditions are met, we consider the table to be an association table and the user interface will take advantage of this fact to streamline various user interactions.
The table may have additional attributes, which can be used to describe additional characteristics of the relationship.